Bulletin of Botanical Research ›› 2025, Vol. 45 ›› Issue (3): 406-418.doi: 10.7525/j.issn.1673-5102.2025.03.011
• Original Paper • Previous Articles Next Articles
Jianhui CHUN1, Wenlong DONG1, Yuanchao TU1, Fang LIU1( ), Yunjian XU2(
), Yunjian XU2( )
)
Received:2024-12-02
Online:2025-05-20
Published:2025-05-23
Contact:
Fang LIU, Yunjian XU
E-mail:liufang0019@ynu.edu.cn;xuyunjian1992@ynu.edu.cn
CLC Number:
Jianhui CHUN, Wenlong DONG, Yuanchao TU, Fang LIU, Yunjian XU. Identification of the Maize GLP Family Genes and Their Expression in Response to Arbuscular Mycorrhizal Symbiosis[J]. Bulletin of Botanical Research, 2025, 45(3): 406-418.
Table 1
Information of ZmGLP gene family members
基因名称 Gene name | 基因ID Gene ID | 氨基酸数目 Number of amino acids/aa | 相对分子量 Relative molecular weight/kDa | 蛋白等电点 Theoretical pI | 不稳定指数 Instability index(Ⅱ) | 脂肪族 氨基酸指数 Aliphatic index | 平均疏水性 Average of hydropathicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| ZmGLP1-1 | Zm00001eb005890 | 230 | 23.96 | 6.04 | 35.85 | 90.87 | 0.20 | 细胞壁 |
| ZmGLP1-2 | Zm00001eb040290 | 114 | 12.52 | 6.26 | 29.95 | 90.70 | 0.08 | 细胞壁 |
| ZmGLP1-3 | Zm00001eb040300 | 232 | 24.55 | 7.01 | 33.53 | 96.68 | 0.18 | 细胞壁 |
| ZmGLP1-4 | Zm00001eb053700 | 225 | 23.79 | 5.89 | 13.41 | 90.89 | 0.08 | 细胞壁 |
| ZmGLP1-5 | Zm00001eb061410 | 228 | 23.72 | 6.04 | 18.95 | 82.98 | 0.14 | 细胞壁 |
| ZmGLP2-1 | Zm00001eb070560 | 263 | 26.90 | 7.80 | 27.42 | 90.84 | 0.28 | 细胞壁 |
| ZmGLP2-2 | Zm00001eb088410 | 226 | 24.60 | 6.40 | 32.27 | 94.51 | 0.05 | 细胞壁 |
| ZmGLP2-3 | Zm00001eb095950 | 208 | 21.65 | 5.34 | 27.25 | 86.44 | 0.32 | 细胞壁 |
| ZmGLP3-1 | Zm00001eb129440 | 218 | 22.82 | 6.57 | 32.98 | 85.55 | 0.20 | 细胞壁 |
| ZmGLP3-2 | Zm00001eb140880 | 208 | 21.34 | 5.59 | 49.81 | 90.58 | 0.13 | 细胞壁 |
| ZmGLP3-3 | Zm00001eb155450 | 220 | 22.95 | 5.88 | 32.98 | 86.95 | 0.33 | 细胞壁 |
| ZmGLP4-1 | Zm00001eb171490 | 157 | 16.92 | 6.25 | 19.75 | 96.24 | 0.09 | 细胞壁 |
| ZmGLP4-2 | Zm00001eb171500 | 228 | 24.81 | 7.77 | 24.61 | 94.12 | 0.03 | 细胞壁 |
| ZmGLP4-3 | Zm00001eb171510 | 228 | 24.80 | 7.77 | 26.54 | 92.81 | 0.02 | 细胞壁 |
| ZmGLP4-4 | Zm00001eb171520 | 228 | 24.64 | 6.50 | 25.51 | 92.41 | 0.03 | 细胞壁 |
| ZmGLP4-5 | Zm00001eb171530 | 229 | 24.77 | 6.28 | 26.18 | 94.59 | 0.05 | 细胞壁 |
| ZmGLP4-6 | Zm00001eb171540 | 229 | 24.79 | 6.28 | 24.75 | 92.88 | 0.02 | 细胞壁 |
| ZmGLP4-7 | Zm00001eb171550 | 228 | 24.64 | 6.89 | 26.12 | 95.00 | 0.08 | 细胞壁 |
| ZmGLP4-8 | Zm00001eb171560 | 127 | 13.89 | 6.96 | 30.13 | 86.06 | -0.13 | 细胞壁 |
| ZmGLP4-9 | Zm00001eb171570 | 228 | 24.61 | 6.50 | 26.55 | 93.29 | 0.05 | 细胞壁 |
| ZmGLP4-10 | Zm00001eb171580 | 229 | 24.78 | 6.57 | 23.89 | 94.15 | 0.06 | 细胞壁 |
| ZmGLP4-11 | Zm00001eb171590 | 209 | 22.78 | 9.21 | 25.83 | 93.78 | -0.06 | 细胞壁 |
| ZmGLP4-12 | Zm00001eb171600 | 228 | 24.55 | 6.27 | 25.00 | 94.12 | 0.05 | 细胞壁 |
| ZmGLP4-13 | Zm00001eb171610 | 228 | 24.57 | 6.57 | 24.79 | 94.12 | 0.05 | 细胞壁 |
| ZmGLP4-14 | Zm00001eb171620 | 193 | 20.94 | 6.30 | 31.98 | 100.98 | -0.04 | 细胞壁 |
| ZmGLP4-15 | Zm00001eb171630 | 229 | 24.79 | 6.57 | 26.11 | 92.88 | 0.04 | 细胞壁 |
| ZmGLP4-16 | Zm00001eb171640 | 228 | 24.72 | 6.96 | 26.92 | 93.29 | 0.03 | 细胞壁 |
| ZmGLP4-17 | Zm00001eb171650 | 228 | 24.52 | 6.50 | 24.21 | 94.56 | 0.07 | 细胞壁 |
| ZmGLP4-18 | Zm00001eb171660 | 222 | 23.82 | 6.89 | 24.34 | 96.26 | 0.12 | 细胞壁 |
| ZmGLP4-19 | Zm00001eb197900 | 257 | 28.02 | 5.83 | 39.10 | 85.37 | 0.24 | 细胞壁 |
| ZmGLP4-20 | Zm00001eb202970 | 218 | 23.25 | 6.88 | 20.79 | 91.74 | 0.27 | 细胞壁 |
| ZmGLP5-1 | Zm00001eb232890 | 224 | 23.09 | 7.74 | 25.23 | 94.20 | 0.31 | 细胞壁 |
| ZmGLP5-2 | Zm00001eb242750 | 234 | 24.58 | 6.52 | 32.21 | 83.08 | 0.06 | 细胞壁 |
| ZmGLP6-1 | Zm00001eb282040 | 212 | 21.89 | 6.01 | 23.72 | 102.12 | 0.53 | 细胞壁 |
| ZmGLP6-2 | Zm00001eb287560 | 227 | 23.25 | 5.11 | 28.34 | 100.22 | 0.49 | 细胞壁 |
| ZmGLP6-3 | Zm00001eb294140 | 215 | 22.44 | 5.89 | 24.18 | 104.33 | 0.44 | 细胞壁 |
| ZmGLP7-1 | Zm00001eb301380 | 231 | 23.57 | 8.58 | 33.12 | 97.66 | 0.33 | 细胞壁 |
| ZmGLP8-1 | Zm00001eb332460 | 214 | 22.36 | 6.57 | 30.74 | 86.68 | 0.18 | 细胞壁 |
| ZmGLP8-2 | Zm00001eb362850 | 226 | 23.38 | 6.89 | 42.54 | 93.76 | 0.22 | 细胞壁 |
| ZmGLP10-1 | Zm00001eb413180 | 226 | 24.63 | 6.18 | 20.73 | 93.19 | -0.05 | 细胞壁 |
| ZmGLP10-2 | Zm00001eb413190 | 226 | 24.69 | 6.18 | 27.33 | 92.74 | 0 | 细胞壁 |
| ZmGLP10-3 | Zm00001eb413210 | 227 | 24.70 | 6.04 | 25.19 | 94.49 | 0 | 细胞壁 |
| ZmGLP10-4 | Zm00001eb413250 | 225 | 24.30 | 6.03 | 25.04 | 94.09 | 0.15 | 细胞壁 |
| ZmGLP10-5 | Zm00001eb413260 | 225 | 24.25 | 6.28 | 29.02 | 94.09 | 0.16 | 细胞壁 |
| ZmGLP10-6 | Zm00001eb419900 | 227 | 24.68 | 6.90 | 31.57 | 94.49 | 0.07 | 细胞壁 |
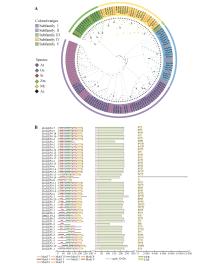
Fig.1
Evolutionary tree of the GLP members(A) and the conserved motifs,protein domains,and gene structures of ZmGLPs(B)A.Evolutionary analysis of GLPs in Zea mays(Zm), Arabidopsis thaliana(At), Oryza sativa(Os), Setaria italica(Si), Medicago truncatula(Mt), and Astragalus sinicus(As); B.Schematic representation of motifs, protein domains, and gene structures of ZmGLP members. From left to right,they represented motifs,protein domains, and gene structures. In the gene structure, green represented UTR(untranslated regions),black lines represented introns,and yellow represented CDS(coding sequences).
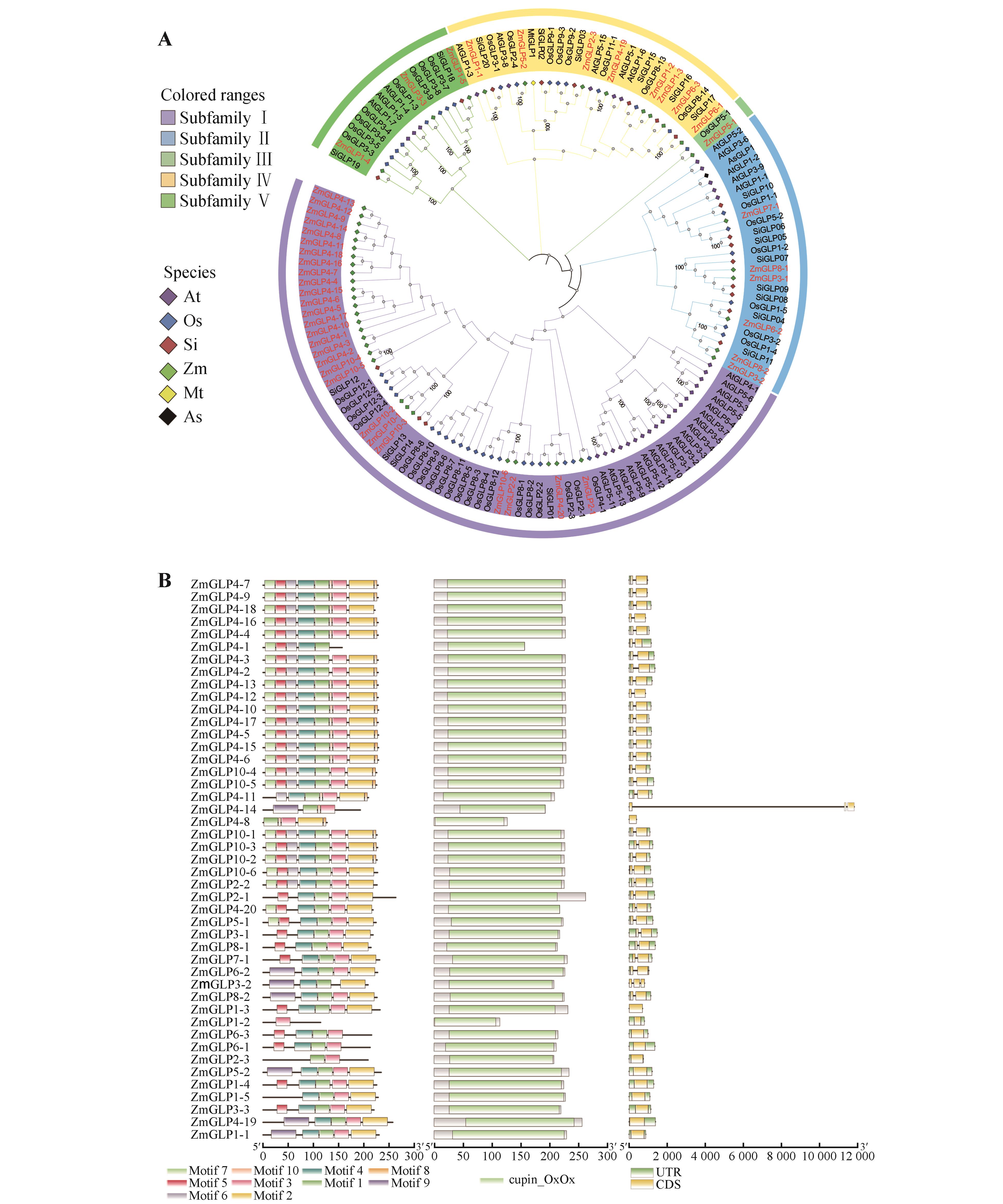
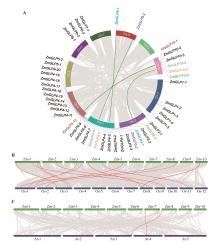
Fig.2
Collinearity analysis of the ZmGLP geneA.Intraspecific collinearity of the ZmGLP genes;B.Collinearity between the ZmGLP genes and the GLP genes in rice;C.Collinearity between the ZmGLP genes and the GLP genes in Arabidopsis thaliana. Green and red lines represented collinear genes,and gene pairs with collinearity were marked with the same color.

Fig.5
Expression patterns of the ZmGLP genes at different times after inoculation with AMF-AMF indicated no inoculation with AMF; +AMF indicated inoculation with AMF; Duncan’s multiple range test, *,**,and *** represented significant differences between treatments at the P<0.05, P<0.01, and P<0.001 levels, respectively.
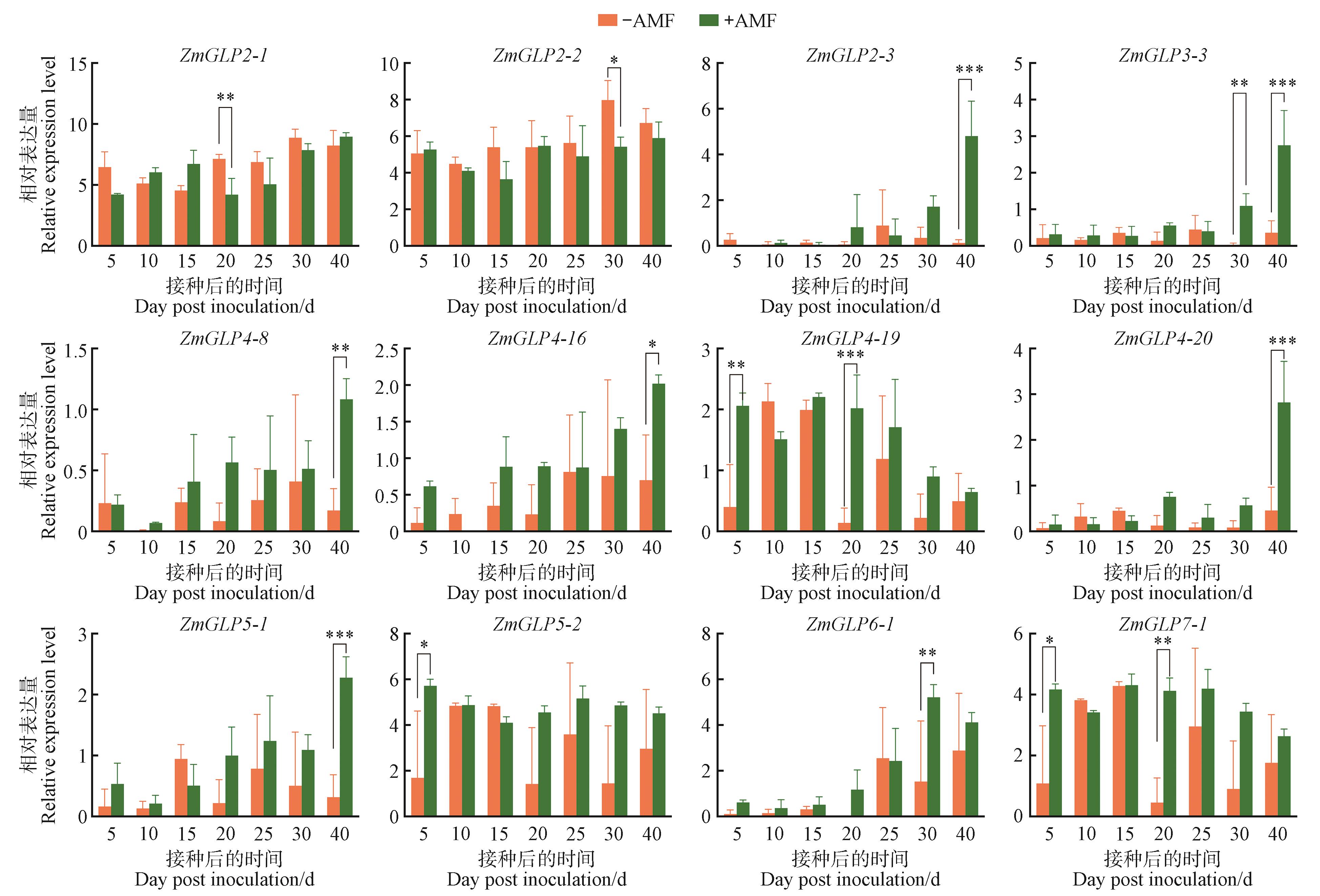
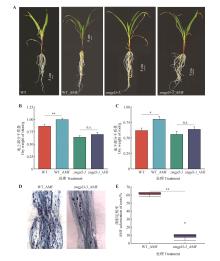
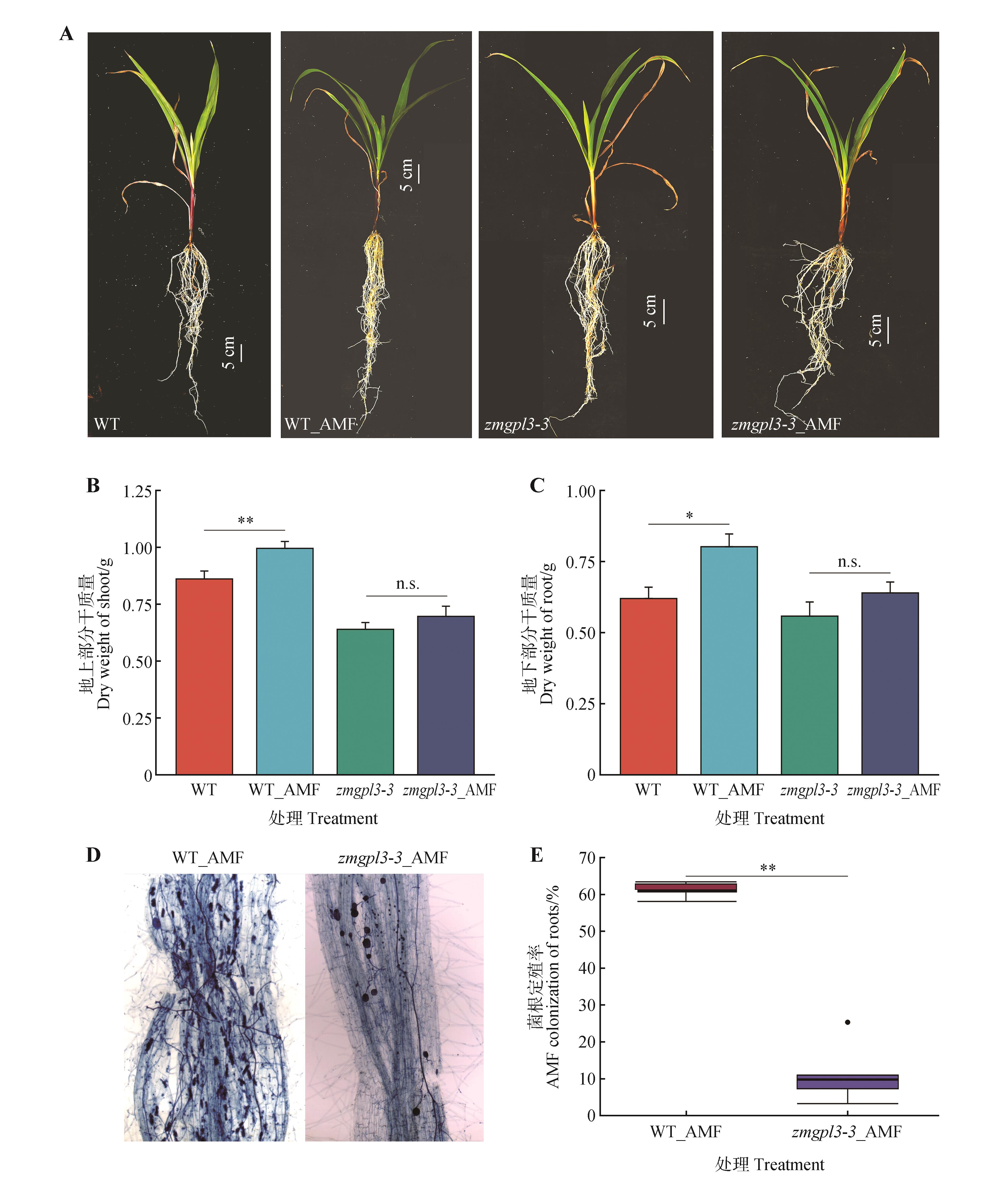
Fig.6
The effect of ZmGLP3-3 mutation on the symbiosis between AMF and maizeA.Phenotypes of maize plants inoculated and non-inoculated with AMF;B.Shoot dry weight of maize plants inoculated and non-inoculated with AMF;C.Root dry weight of maize plants inoculated and non-inoculated with AMF;D.Trypan blue staining of maize mycorrhizae after inoculation with AMF;E.Mycorrhizal symbiosis rate. AMF indicated inoculation with AMF; WT represented the wild-type maize plants, zmgpl3-3 represented the mutant; * and ** indicated significant differences at P<0.05 and P<0.01 levels, respectively; n.s. indicated no significant difference.
| 1 | SMITH S E, SMITH F A.Roles of arbuscular mycorrhizas in plant nutrition and growth:new paradigms from cellular to ecosystem scales[J].Annual Review of Plant Biology,2011,62(1):227-250. |
| 2 | LIU C Y, GUO X N, DAI F J,et al.Mycorrhizal symbiosis enhances P uptake and indole-3-acetic acid accumulation to improve root morphology in different citrus genotypes[J].Horticulturae,2024,10(4):339. |
| 3 | FU W, CHEN B D, RILLIG M C,et al.Community response of arbuscular mycorrhizal fungi to extreme drought in a cold-temperate grassland[J].New Phytologist,2022,234(6):2003-2017. |
| 4 | 黄静娴.植物-丛枝菌根真菌共生的研究进展[J].世界生态学,2024,13(2):255-261. |
| HUANG J X.Research progress on plant-arbuscular mycorrhizal fungi symbiosis[J].International Journal of Ecology,2024,13(2):255-261. | |
| 5 | SHI J C, WANG X L, WANG E T.Mycorrhizal symbiosis in plant growth and stress adaptation:from genes to ecosystems[J].Annual Review of Plant Biology,2023,74(1):569-607. |
| 6 | DUAN S L, FENG G, LIMPENS E,et al.Cross-kingdom nutrient exchange in the plant-arbuscular mycorrhizal fungus-bacterium continuum[J].Nature Reviews Microbiology,2024,22(12):773-790. |
| 7 | DUNWELL J M, PURVIS A, KHURI S.Cupins:the most functionally diverse protein superfamily?[J].Phytochemistry,2004,65(1):7-17. |
| 8 | DUNWELL J M, GIBBINGS J G, MAHMOOD T,et al.Germin and germin-like proteins:evolution,structure,and function[J].Critical Reviews in Plant Sciences,2008,27(5):342-375. |
| 9 | AGARWAL G, RAJAVEL M, GOPAL B,et al.Structure-based phylogeny as a diagnostic for functional characterization of proteins with a cupin fold[J].PLoS One,2009, 4(5):e5736. |
| 10 | BARMAN A R, BANERJEE J.Versatility of germin-like proteins in their sequences,expressions,and functions[J].Functional & Integrative Genomics,2015,15(5):533-548. |
| 11 | GOVINDAN G, SANDHIYA K R, ALPHONSE V,et al.Role of germin-like proteins(GLPs) in biotic and abiotic stress responses in major crops:a review on plant defense mechanisms and stress tolerance[J].Plant Molecular Biology Reporter,2024,42(3):450-468. |
| 12 | PEI Y K, LI X C, ZHU Y T,et al.GhABP19,a novel germin-like protein from Gossypium hirsutum,plays an important role in the regulation of resistance to Verticillium and Fusarium wilt pathogens[J].Frontiers in Plant Science,2019,10:583. |
| 13 | MAO L X, GE L J, YE X C,et al.ZmGLP1,a germin-like protein from maize,plays an important role in the regulation of pathogen resistance[J].International Journal of Molecular Sciences,2022,23(22):14316. |
| 14 | YUAN B J, YANG Y L, FAN P,et al.Genome-wide identification and characterization of germin and germin-like proteins(GLPs) and their response under powdery mildew stress in wheat (Triticum aestivum L.)[J].Plant Molecular Biology Reporter,2021,39(4):821-832. |
| 15 | ZHANG Y H, WANG X S, CHANG X C,et al.Overexpression of germin-like protein GmGLP10 enhances resistance to Sclerotinia sclerotiorum in transgenic tobacco[J].Biochemical and Biophysical Research Communications,2018,497(1):160-166. |
| 16 | GUCCIARDO S, WISNIEWSKI J P, BREWIN N J,et al.A germin-like protein with superoxide dismutase activity in pea nodules with high protein sequence identity to a putative rhicadhesin receptor[J].Journal of Experimental Botany,2007,58(5):1161-1171. |
| 17 | DOLL J, HAUSE B, DEMCHENKO K,et al.A member of the germin-like protein family is a highly conserved mycorrhiza-specific induced gene[J].Plant and Cell Physiology,2003,44(11):1208-1214. |
| 18 | ZENG X B, LI D Z, LV Y F,et al.A germin-like protein GLP1 of legumes mediates symbiotic nodulation by interacting with an outer membrane protein of rhizobia[J].Microbiology Spectrum,2023,11(1):e0335022. |
| 19 | PAWLOWSKI M L, VUONG T D, VALLIYODAN B,et al.Whole-genome resequencing identifies quantitative trait loci associated with mycorrhizal colonization of soybean[J].Theoretical and Applied Genetics,2020,133:409-417. |
| 20 | ZORIN E A, SULIMA A S, ZHERNAKOV A I,et al.Genomic and transcriptomic analysis of pea(Pisum sativum L.) breeding line ‘Triumph’ with high symbiotic responsivity[J].Plants,2024,13(1):78. |
| 21 | HE H X.Study on the current situation and influencing factors of corn import trade in China:based on the trade gravity model[J].Journal of Intelligent Systems,2024, 33(1):20240040. |
| 22 | HUI J, AN X, LI Z B,et al.The mycorrhiza-specific ammonium transporter ZmAMT3;1 mediates mycorrhiza-dependent nitrogen uptake in maize roots[J].The Plant Cell,2022,34(10):4066-4087. |
| 23 | ILYAS M,ALI I, NASSER BINJAWHAR D,et al.Molecular characterization of germin-like protein genes in Zea mays (ZmGLPs) using various in silico approaches[J].ACS Omega,2023,8(18):16327-16344. |
| 24 | LI L, XU X H, CHEN C,et al.Genome-wide characterization and expression analysis of the germin-like protein family in rice and Arabidopsis [J].International Journal of Molecular Sciences,2016,17(10):1622. |
| 25 | CHEN C J, WU Y, LI J W,et al.TBtools-II:a “one for all,all for one” bioinformatics platform for biological big-data mining[J].Molecular Plant,2023,16(11):1733-1742. |
| 26 | BILESCHI M L, BELANGER D, BRYANT D H,et al.Using deep learning to annotate the protein universe[J].Nature Biotechnology,2022,40:932-937. |
| 27 | LU S N, WANG J Y, CHITSAZ F,et al.CDD/SPARCLE:the conserved domain database in 2020[J].Nucleic Acids Research,2020,48(D1):265-268. |
| 28 | CHOU K C, SHEN H B.Plant-mPLoc:a top-down strategy to augment the power for predicting plant protein subcellular localization[J].PLoS One,2010,5(6):e11335. |
| 29 | SAHA D, RANA R S, ARYA L,et al.Genomic organization and structural diversity of germin-like protein coding genes in foxtail millet (Setaria italica L.)[J].Agri Gene,2017,3:87-98. |
| 30 | LETUNIC I, BORK P.Interactive Tree of Life(iTOL) v6:recent updates to the phylogenetic tree display and annotation tool[J].Nucleic Acids Research,2024,52(W1):78-82. |
| 31 | WANG Y P, TANG H B, WANG X Y,et al.Detection of colinear blocks and synteny and evolutionary analyses based on utilization of MCScanX[J].Nature Protocols,2024,19(7):2206-2229. |
| 32 | WANG Y Z, JIA L H, TIAN G,et al.shinyCircos-V2.0:leveraging the creation of circos plot with enhanced usability and advanced features[J].iMeta,2023,2(2):e109. |
| 33 | LESCOT M, DÉHAIS P, THIJS G,et al.PlantCARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J].Nucleic Acids Research,2002,30(1):325-327. |
| 34 | WANG X B, ZHANG H W, GAO Y L,et al.A comprehensive analysis of the Cupin gene family in soybean (Glycine max)[J].PLoS One,2014,9(10):e110092. |
| 35 | DRUKA A, KUDRNA D, KANNANGARA C G,et al.Physical and genetic mapping of barley (Hordeum vulgare) germin-like cDNAs[J].Proceedings of the National Academy of Sciences of the United States of America,2002,99(2):850-855. |
| 36 | ZHANG Z D, WEN Y S, YUAN L Q,et al.Genome-wide identification,characterization,and expression analysis related to low-temperature stress of the CmGLP gene family in Cucumis melo L.[J].International Journal of Molecular Sciences,2022,23(15):8190. |
| 37 | SANG Y M, LIU Q F, LEE J,et al.Expansion of amphibian intronless interferons revises the paradigm for interferon evolution and functional diversity[J].Scientific Reports,2016,6(1):29072. |
| 38 | ILYAS M, RASHEED A, MAHMOOD T.Functional characterization of germin and germin-like protein genes in various plant species using transgenic approaches[J].Biotechnology Letters,2016,38(9):1405-1421. |
| 39 | CHEN A Q, GU M, SUN S B,et al.Identification of two conserved cis-acting elements,MYCS and P1BS,involved in the regulation of mycorrhiza-activated phosphate transporters in eudicot species[J].New Phytologist,2011,189(4):1157-1169. |
| [1] | Wenhui YAN, Shilong DUAN, Lin ZHANG. Mechanisms and Regulation of Interactions between AM Fungi and Hyphosphere Bacteria in Organic Phosphorus Mineralization [J]. Bulletin of Botanical Research, 2025, 45(3): 345-351. |
| [2] | Boyan WANG, Jin CHEN, Qixiu CHENG, Yueming BAO, Haining WANG, Rui QIN, Xiaoyu LI. Progress of Research on Culture and Application of Arbuscular Mycorrhizal Fungi [J]. Bulletin of Botanical Research, 2025, 45(3): 361-370. |
| [3] | Wu LIU, Yaying YANG, Ning GONG, Ziwei ZOU, Yi WANG, Baodong CHEN, Qiong WANG, Wei LIU. Growth and Physiological Responses of Ancient Celtis sinensis Seedlings to the Inoculation of Indigenous Arbuscular Mycorrhizal Fungi [J]. Bulletin of Botanical Research, 2025, 45(3): 393-405. |
| [4] | Baoru XUN, Hongtao QIN, Rui MA, Nanfeng GUO, Yunping LIU, Ying WU, Xingguo LAN. Gene Cloning, Expression and Interaction Protein Analysis of FERONIA in Brassica oleracea var. acephala [J]. Bulletin of Botanical Research, 2024, 44(2): 298-306. |
| [5] | Chunyao WANG, Xiaojin LEI, Zhongyuan LIU. Expression Pattern Analysis of PdbHMGs Genes in Populus davidiana×P. bolleana under Abiotic Stress [J]. Bulletin of Botanical Research, 2023, 43(6): 932-942. |
| [6] | Anying HUANG, Dean XIA, Yang ZHANG, Dongchen NA, Qing YAN, Zhigang WEI. Cloning and Drought Tolerance Expression Analysis of PtrWRKY51 Gene in Populus trichocarpa [J]. Bulletin of Botanical Research, 2022, 42(6): 1005-1013. |
| [7] | Huafeng CHEN, Longjun DAI, Mingyang LIU, Bingbing GUO, Hong YANG, Lifeng WANG. Stress Tolerance Functional Analysis of the High Expression Heat Shock Protein HbHSP90.4 Gene from the Latex of Hevea brasiliensis [J]. Bulletin of Botanical Research, 2022, 42(6): 1023-1032. |
| [8] | Mingyang LIU, Huaxing XIAO, Lifeng WANG, Xiaoxu LIANG, Yu ZHANG, Meng WANG. Cloning and Functional Analysis of Heat Shock Protein HbHSP90.8-1 from Hevea brasiliensis Müll. Arg. [J]. Bulletin of Botanical Research, 2022, 42(5): 811-820. |
| [9] | Zhaoyi LI, Longfei HAO, Tingyan LIU, Yanhong HE, You ZHANG, Shulan BAI, Xinyu YANG. AM Fungi Inoculation on Root Morphology and Nutrient Loading of Clematis fruticosa Seedlings under Simulated Atmospheric Nitrogen Deposition [J]. Bulletin of Botanical Research, 2022, 42(5): 886-895. |
| [10] | Xueying WANG, Ruiqi WANG, Yang ZHANG, Cong LIU, Dean XIA, Zhigang WEI. Genome‑wide Identification and Stress Response Analysis of Cyclic Nucleotide-gated Channels(CNGC) Gene Family in Populus trichocarpa [J]. Bulletin of Botanical Research, 2022, 42(4): 613-625. |
| [11] | Yuning Yang, Hao Dong, Shiwei Dong, Nairui Wang, Yue Song, Hanguo Zhang, Shujuan Li. Cloning and Expression Analysis of Transcription Factor LobHLH34 from Larix olgensis [J]. Bulletin of Botanical Research, 2022, 42(1): 112-120. |
| [12] | Shuang-Hui TIAN, He CHENG, Yang ZHANG, Cong LIU, De-An XIA, Zhi-Gang WEI. Genome-wide Identification and Expressional Analysis of Carotenoid Cleavage Dioxygenases(CCD) Gene Family in Populus trichocarpa under Drought and Salt Stress [J]. Bulletin of Botanical Research, 2021, 41(6): 993-1005. |
| [13] | Xin-Tong JI, Lei YU, Ya-Guang ZHAN. Cloning and Expression Analysis of BRASSINAZOLE RESISTANTANT1 (BZR1) Gene from Fraxinus mandshurica [J]. Bulletin of Botanical Research, 2021, 41(5): 744-752. |
| [14] | Shu-Ping PENG, Cheng-Ming DONG, Yun-Hao ZHU. Cloning and Expression Analysis of Two Key Genes of Jasmonic Acid Synthesis in Response to Endophytic Infection from Rehmannia glutinosa [J]. Bulletin of Botanical Research, 2021, 41(2): 294-301. |
| [15] | Xiao-Xiao WANG, Bi QIN, Yu-Shuang YANG, Qiu-Hai NIE, Ji-Chuan ZHANG, Shi-Zhong LIU. Cloning and Expression Analysis of E2 Ubiquitin-conjugating Enzyme Gene TkUBC2 in Taraxacum kok-saghyz Rodin [J]. Bulletin of Botanical Research, 2021, 41(1): 98-106. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||