Bulletin of Botanical Research ›› 2023, Vol. 43 ›› Issue (3): 340-350.doi: 10.7525/j.issn.1673-5102.2023.03.003
• Genetic and Breeding • Previous Articles Next Articles
Zhanmin ZHENG1, Yubing SHANG1, Guangbo ZHOU1, Di XIAO2,3, Yi LIU3, Xiangling YOU4( )
)
Received:2022-10-03
Online:2023-05-20
Published:2023-06-06
Contact:
Xiangling YOU
E-mail:youxiangling@nefu.edu.cn
About author:E-mail:youxiangling@nefu.edu.cnSupported by:CLC Number:
Zhanmin ZHENG, Yubing SHANG, Guangbo ZHOU, Di XIAO, Yi LIU, Xiangling YOU. Genetic Transformation and Function Analysis of PsnHB13 and PsnHB15 of Populus simonii × Populus nigra[J]. Bulletin of Botanical Research, 2023, 43(3): 340-350.
Table 1
Primers used in this experiment
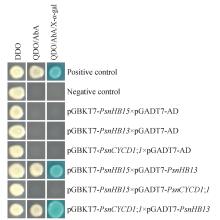
Fig.4
Yeast two-hybrid interaction for verificationDDO.SD/-Trp/-Leu medium;QDO/A.SD/-Trp/-Leu/-His/-Ade/AbA medium;QDO/X/A.SD/-Trp/-Leu/-His/-Ade/X-α-gal/AbA medium;Positive control. pGBKT7-53 and pGADT7-T plasmid co-transformation;Negative control.pGBKT7-Lam and pGADT7-T plasmid co-transformation
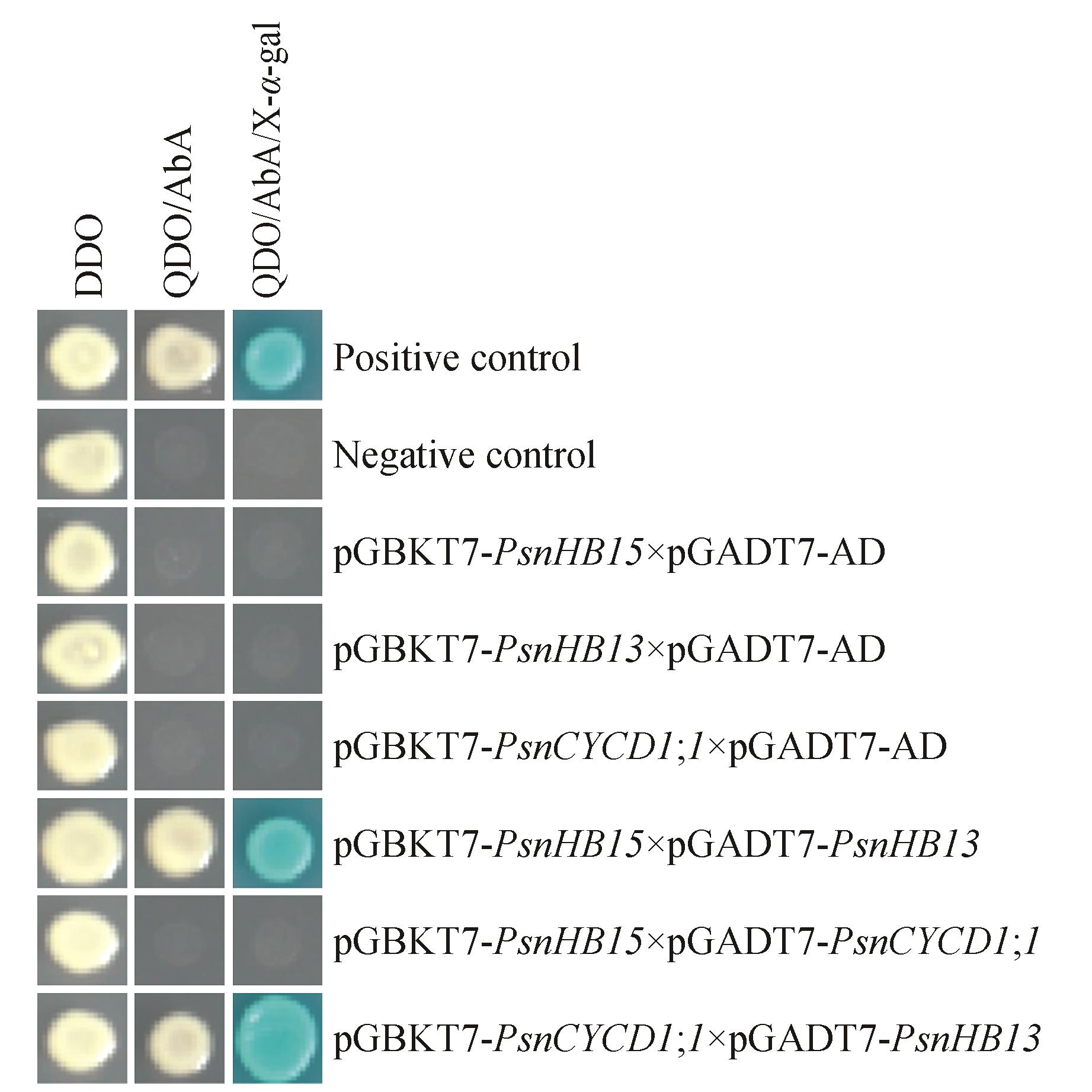
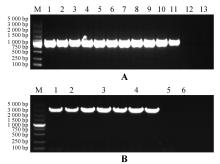
Fig.5
DNA detection of transgenic strainsA.PCR detection electrophoresis of PsnHB13 overexpression transgenic lines DNA(1.Positive control;2-11.10 transgenic lines;12.Wild-type plant control;13.ddH2O as negative control);B.PCR detection electrophoresis of PsnHB15 overexpression transgenic lines DNA(1.positive control;2-4. 3 transgenic poplar lines;5.Wild type plant control;6.ddH2O as negative control);M.DL5000 Marker
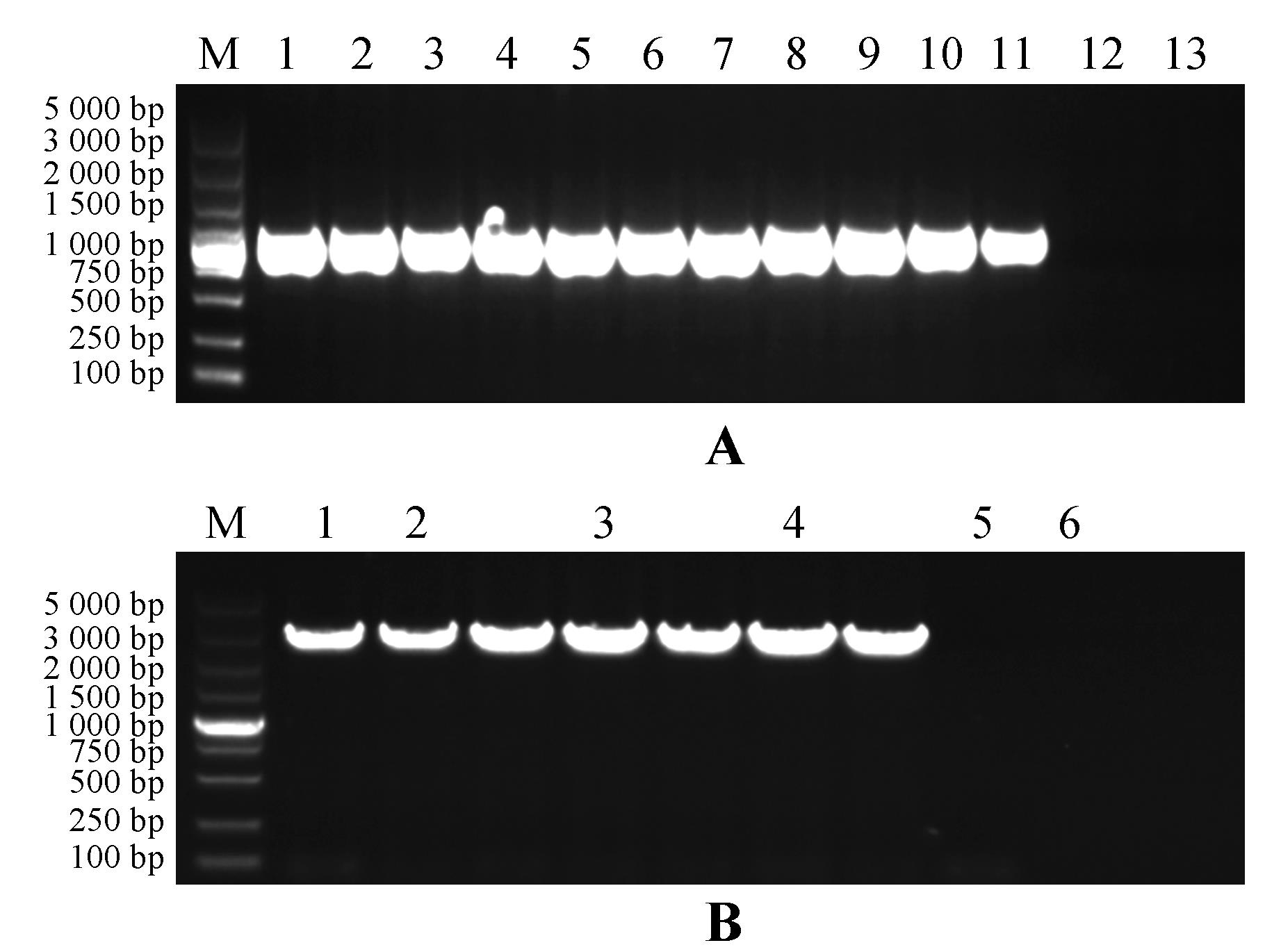
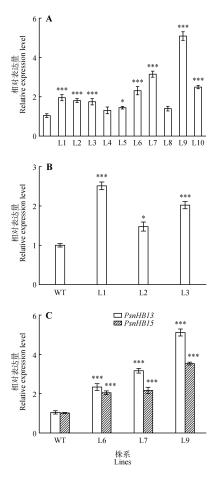
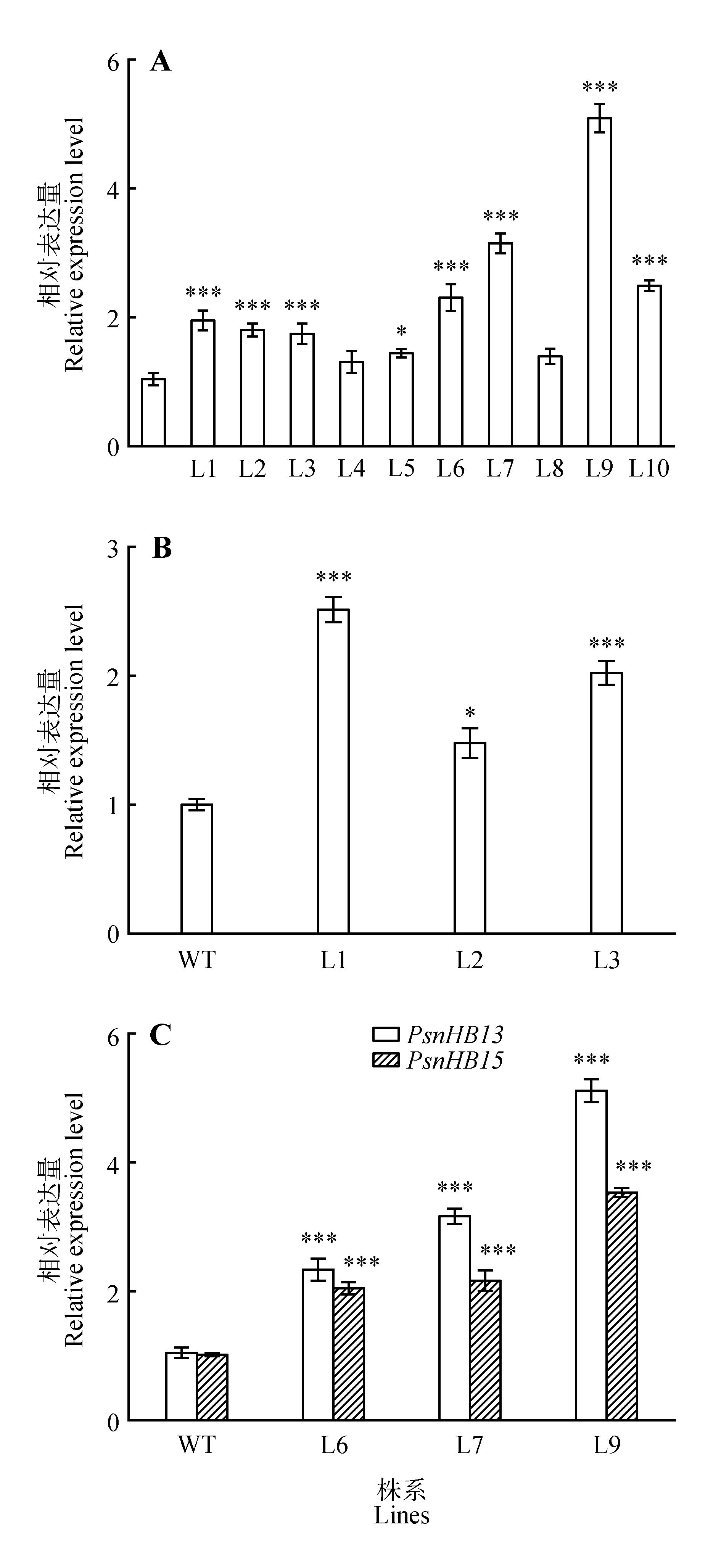
Fig.6
Detection of the relative expression of transgenic linesA.PsnHB13 gene expression in different PsnHB13 overexpression lines(WT.Wild type;L1-L10.Different transgenic lines);B.PsnHB15 gene expression in different PsnHB15 overexpression lines(WT.Wild type;L1-L3.Different transgenic lines);C.PsnHB15 gene expression in differentPsnHB13 overexpression lines(WT.Wild type;L6,L7,L9.Different transgenic lines)
| 1 | MENGES M, DE JAGER S M, GRUISSEM W,et al.Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes,reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control[J].The Plant Journal,2005,41(4):546-566. |
| 2 | SORRELL D A, COMBETTES B, CHAUBET-GIGOT N,et al.Distinct Cyclin D genes show mitotic accumulation or constant levels of transcripts in tobacco bright yellow-2 cells[J].Plant physiology(Bethesda),1999,119(1):343-352. |
| 3 | DE VEYLDER L.The discovery of plant D-Type cyclins[J].The Plant Cell,2019,31(6):1194-1195. |
| 4 | HU Y, BAO F, LI J.Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis[J].The Plant Journal,2000,24(5):693-701. |
| 5 | RIOU-KHAMLICHI C, MENGES M, HEALY J M,et al.Sugar control of the plant cell cycle:differential regulation of Arabidopsis D-type cyclin gene expression[J].Molecular and Cellular Biology,2000,20(13):4513-4521. |
| 6 | ZHENG T C, DAI L J, LIU Y,et al.Overexpression Populus d-type cyclin gene PsnCYCD1;1 influences cell division and produces curved leaf in Arabidopsis thaliana [J].International Journal of Molecular Sciences,2021,22(11):5837. |
| 7 | 郑唐春.小黑杨PsnCYCD1;1基因参与细胞分裂及次生生长作用机理研究[D].哈尔滨:东北林业大学,2016. |
| ZHENG T C.Study on mechanism of Populus simonii × P .nigra PsnCYCD 1;1 gene involved in cell division and secondary growth[D].Harbin:Northeast Forestry University,2016. | |
| 8 | VIOLA I L, GONZALEZ D H.Chapter 6-structure and evolution of plant homeobox genes[M]//GONZALEZ D H.Plant transcription factors:evolutionary,structural and functional aspects.New York:Academic Press,2016:101-112. |
| 9 | 李俐,王义,王康宇,等. HD-Zip基因家族的结构、功能与表达模式[J].分子植物育种.2018,16(3):781-790. |
| LI L, WANG Y, WANG K Y,et al.The structure,function and expression pattern of HD-Zip gene family[J].Molecular Plant Breeding,2018,16(3):781-790. | |
| 10 | 卢小云,余礼,张秀峰,等.禾本科植物HD-Zip转录因子研究进展[J].植物生理学报,2021,57(4):727-738. |
| LU X Y, YU L, ZHANG X F. et al.Research progress of HD-Zip transcription factor in gramineae plants[J].Plant Physiology Journal,2021,57(4):727-738. | |
| 11 | CAPELLA M.Chapter 7-Homeodomain-Leucine Zipper transcription factors:structural features of these proteins,unique to plants[M]//RIBONE P A,ARCE A L,CHAN R L,et al.Plant transcription factors:evolutionary,structural and functional aspects.New York:Academic Press,2016:113-126. |
| 12 | ARCE A L, RAINERI J, CAPELLA M,et al.Uncharacterized conserved motifs outside the HD-Zip domain in HD-Zip subfamily Ⅰ transcription factors; a potential source of functional diversity[J].BMC Plant Biology,2011,11:42. |
| 13 | 王家啟,张曦,李莉.白桦HD-Zip基因家族生物信息学及应答盐胁迫分析[J].植物研究.2018,38(6):931-938. |
| WANG J Q, ZHANG X, LI L.Bioinformatic and expression analysis of HD-Zip family gene in Betula platyphylla [J].Bulletin of Botanical Research,2018,38(6):931-938. | |
| 14 | HENRIKSSON E, OLSSON A S B, JOHANNESSON H,et al.Homeodomain leucine zipper class Ⅰ genes in Arabidopsis.Expression patterns and phylogenetic relationships[J].Plant Physiology,2005,139(1):509-518. |
| 15 | AOYAMA T, DONG C H, WU Y,et al.Ectopic expression of the Arabidopsis transcriptional activator Athb-1 alters leaf cell fate in tobacco[J].The Plant Cell,1995,7(11):1773-1785. |
| 16 | KIM Y K,SON O, KIM M R,et al. ATHB23,an Arabidopsis class Ⅰ homeodomain-leucine zipper gene,is expressed in the adaxial region of young leaves[J].Plant Cell Reports,2007,26(8):1179-1185. |
| 17 | 肖迪,刘轶,李开隆,等.小黑杨PsnHB13基因在烟草中的遗传转化与功能分析[J].植物研究,2020,40(4):593-601. |
| XIAO D, LIU Y, LI K L,et al.Genetic transformation and function analysis of PsnHB13 gene isolated from Populus simonii × P. nigra in Nicotiana tabacum [J].Bulletin of Botanical Research,2020,40(4):593-601. | |
| 18 | HANSON J, REGAN S, ENGSTRÖM P.The expression pattern of the homeobox gene ATHB13 reveals a conservation of transcriptional regulatory mechanisms between Arabidopsis and hybrid aspen[J].Plant Cell Reports,2002,21(1):81-89. |
| 19 | MUKHERJEE K, BÜRGLIN T R.MEKHLA,a novel domain with similarity to PAS domains,is fused to plant homeodomain-leucine zipper Ⅲ proteins[J].Plant Physiology,2006,140(4):1142-1150. |
| 20 | ROMANOWSKI M J, SOCCIO R E, BRESLOW J L,et al.Crystal structure of the Mus musculus cholesterol-regulated START protein 4(StarD4) containing a StAR-related lipid transfer domain[J].Proceedings of the National Academy of Sciences of the United States of America,2002,99(10):6949-6954. |
| 21 | SAKAKIBARA K, NISHIYAMA T, KATO M,et al.Isolation of homeodomain-leucine zipper genes from the moss Physcomitrella patens and the evolution of homeodomain-leucine zipper genes in land plants[J].Molecular Biology and Evolution,2001,18(4):491-502. |
| 22 | OTSUGA D, DEGUZMAN B, PRIGGE M J,et al.REVOLUTA regulates meristem initiation at lateral positions[J].The Plant Journal,2001,25(2):223-236. |
| 23 | AGALOU A, PURWANTOMO S, ÖVERNÄS E,et al.A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members[J].Plant Molecular Biology,2008,66(1-2):87-103. |
| 24 | PRIGGE M J, OTSUGA D, ALONSO J M,et al.Class Ⅲ homeodomain-leucine zipper gene family members have overlapping,antagonistic,and distinct roles in Arabidopsis development[J].The Plant Cell,2005,17(1):61-76. |
| 25 | BRANDT R, XIE Y K, MUSIELAK T,et al.Control of stem cell homeostasis via interlocking microRNA and microProtein feedback loops[J].Mechanisms of Development,2013,130(1):25-33. |
| 26 | OHASHI-ITO K, FUKUDA H.HD-Zip III homeobox genes that include a novel member,ZeHB-13(Zinnia)/ATHB-15(Arabidopsis),are involved in procambium and xylem Cell differentiation[J].Plant & Cell Physiology,2003,44(12):1350-1358. |
| 27 | TANG G L, REINHART B J, BARTEL D P,et al.A biochemical framework for RNA silencing in plants[J].Genes & Development,2003,17(1):49-63. |
| 28 | RHOADES M W, REINHART B J, LIM L P,et al.Prediction of plant microRNA targets[J].Cell,2002,110(4):513-520. |
| 29 | ZHU Y Y, SONG D L, XU P,et al.A HD-ZIP Ⅲ gene,PtrHB4,is required for interfascicular cambium development in Populus [J].Plant Biotechnology Journal,2018,16(3):808-817. |
| 30 | ZHU Y Y, SONG D L, SUN J Y,et al. PtrHB7,a class ⅢHD-Zip gene,plays a critical role in regulation of vascular cambium differentiation in Populus [J].Molecular Plant,2013,6(4):1331-1343. |
| [1] | Senyao LIU, Fenglin JIA, Qing GUO, Gaofeng FAN, Boru ZHOU, Tingbo JIANG. Response Analysis of Transcription Factor PsnbHLH162 Gene in Populus simonii × P. nigra under Salt Stress and Low Temperature Stress [J]. Bulletin of Botanical Research, 2023, 43(2): 300-310. |
| [2] | Jingya Yu, Mingze Xia, Hao Xu, Faqi Zhang. Comparative Transcriptome Analysis of Three Artemisia Species in Qinghai Tibet Plateau [J]. Bulletin of Botanical Research, 2022, 42(2): 200-210. |
| [3] | YE Xing-Zhuang, LIU Dan, LUO Jia-Jia, FAN Hui-Hua, ZHANG Guo-Fang, LIU Bao, CHEN Shi-Pin. Transcriptome Analysis for Rare and Endangered Plants of Semiliquidambar cathayensis [J]. Bulletin of Botanical Research, 2019, 39(2): 276-286. |
| [4] | HONG Sen-Rong, WU Hui, ZHONG Lu, XIONG Si-Min, LUO Xia, LIU Xing. Transcriptome Analysis of Dioscorea bulbifera L. Microtubers Conserved in vitro at Low Temperature [J]. Bulletin of Botanical Research, 2018, 38(1): 100-109. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||